Chemistry
Computation in Chemistry
Historically, chemistry was understood as being organized into two broad categories: experimental and computational. However, that classification system has become increasingly inadequate to describe the fascinating and diverse types of chemistry research now available due to increased computing capabilities. For example, computational models can help us estimate how molecules fit into enzymes to control their expression, so we can maximize the time and materials devoted to drug discovery. Likewise, innovative methodology like leaf spray ionization rapidly develops massive data sets as we endeavor to understand plants’ intricate response to predators. Furthermore, harnessing the computing power of supercomputers enables physical chemists to elucidate the vibrational modes in crystal structures. Therefore, for these and many other chemistry applications, it is vital to develop the knowledge, skills, and experience for students to learn computations skills to work on modern, complex chemical studies.
 Phil Wittel presenting at Eli Lilly & Company</span
Phil Wittel presenting at Eli Lilly & Company</span
 Phil Wittel presenting at the Butler Undergraduate Research Conference
Phil Wittel presenting at the Butler Undergraduate Research Conference
Questions addressed by computers in chemistry
How do intermolecular forces determine how small molecules with various side chains fit into the active site of an enzyme or DNA?
How does the age of a plant and the location of a leaf affect how rapidly can a plant produce and expel molecules to deter predators from consuming them?
Computation in Chemistry and Biochemistry at UE
The curriculum in the Department of Chemistry at UE is designed to prepare students for a variety of satisfying and meaningful careers. Our students begin contributing to the scientific community as undergraduate researchers, then continue on to careers in industry, graduate degrees, or professional schools. Our dedicated faculty mentor students in undergraduate research in a diverse variety of computational-based projects.
The Lampkins Supramolecular Chemistry Lab uses a program called Autodock Vina to virtually test compounds that we design to inhibit the replication of gram-negative, drug resistant bacteria.
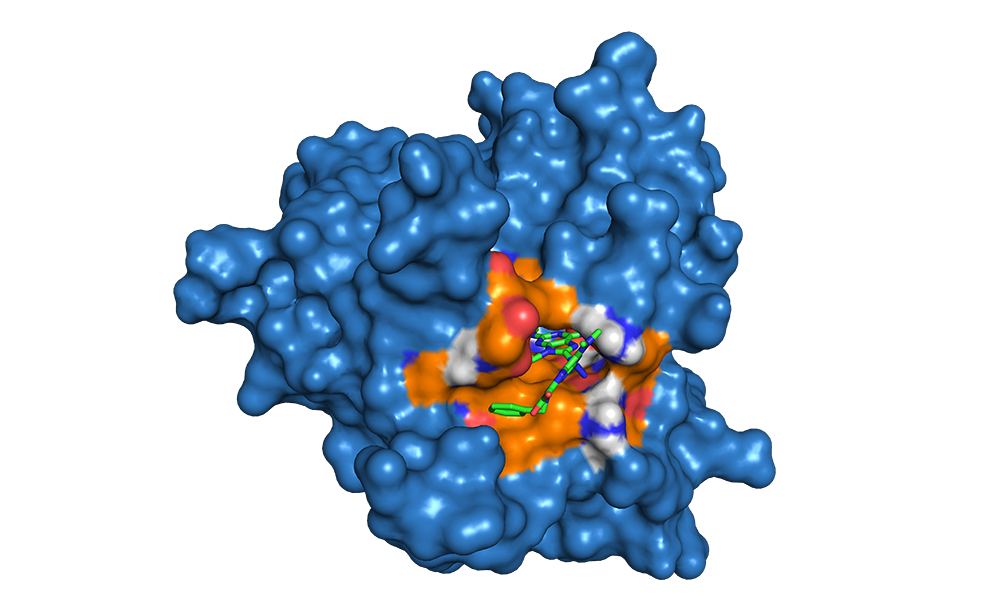
Target compound in the ATP-binding site of E. Coli gyrase B using Autodock Vina
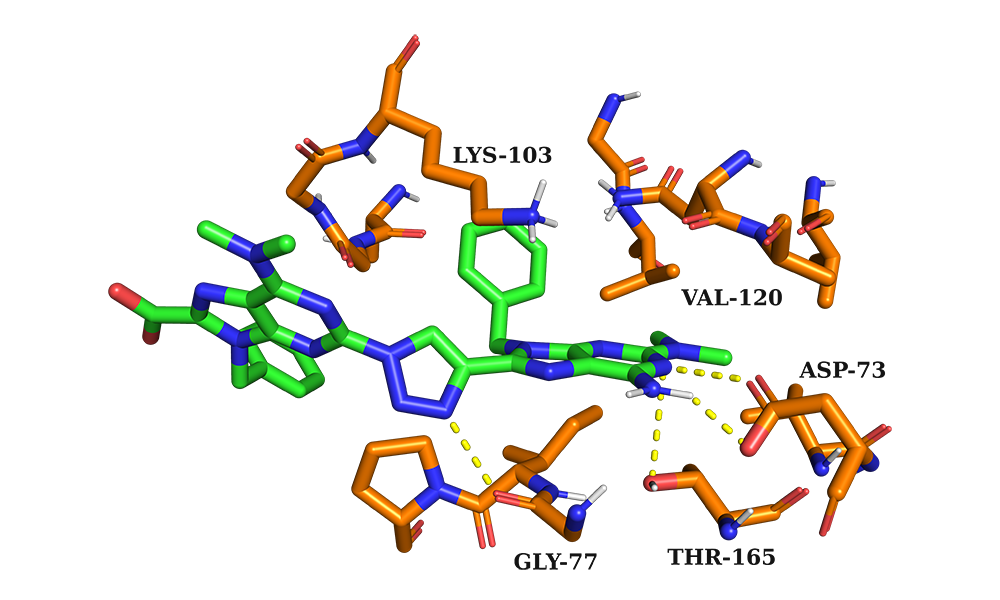
The binding mode of the target compound in the ATP binding site of E.Coli gyrase B using Autodock Vina
Dr. Lampkins obtained grant funding and mentored Phil Wittel, who performed a computational chemistry visualization project in which he modeled potential target molecules for their fit in the ATP binding site of E. Coli gyrase B in AutoDock Vina. The results then informed the selection of molecule targets for organic chemistry undergraduate research also being pursued in the group.
Dr. Lynch obtained funding from the National Science Foundation (NSF) XSEDE program to utilize time on the supercomputer Stampede to study vibrations in crystals of cyclopentane.
Office Phone
812-488-2029
Office Email
cism@evansville.edu
Office Location
Room 216, Koch Center for Engineering and Science
